Abstract
Background: Survival rates for pediatric acute myeloid leukemia (AML) still lag far behind those for most other pediatric cancers. New agents are desperately needed, but new drug development has been hampered by a lack of clinically relevant pediatric AML patient-derived xenograft (PDX) models for research. Texas Children's Hospital (TCH) has the patient volume, a close collaboration between clinical groups and research laboratories, and the infrastructure to address this gap.
Methods: TCH patients diagnosed with AML are offered participation in local research. Most samples from local patients are enriched for mononuclear cells (MNCs) and injected fresh on the day of collection. Additionally, cryopreserved bone marrow samples from patients who enrolled on Children's Oncology Group clinical trials and consented to bank tissue for research have been used for PDX generation. Leukemia cells are tail vein injected (2x10e5 cells per mouse) into immunodeficient mice without conditioning. Most models are generated in the NOD.scid.Il2Rγc-/-/SCFh/h/GM-CSFh/h/IL-3h/h (NSGS) strain. Mice are closely monitored for human CD45+/CD33+/CD3- (hAML) cells in peripheral blood by flow cytometry. Once moribund, the mice are humanely euthanized, and bone marrow, peripheral blood, and spleen are harvested and evaluated by flow cytometry for disease burden. Bone marrow cells are injected by tail vein into secondary recipients. Serially transplanting PDX models are validated and characterized by short tandem repeat (STR) fingerprinting and DNA/RNA sequencing.
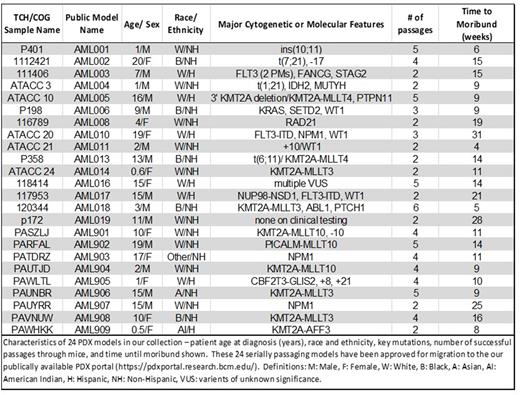
Results: Thus far, we have developed 24 serially transplanting pediatric AML PDX models representing a variety of clinical and cytomolecular categories. The patients from whom these models were derived ranged in age from 0.5-20 years of age at diagnosis (mean 9.4 years). Most are derived from samples collected at diagnosis. For AML005, we have a second serially transplanting model derived from relapse. Our collection includes 8 models with KMT2A fusions, 3 with NPM1 mutations, 4 with WT1 mutations, 3 with FLT3 internal tandem duplications or point mutations, 1 with a NUP98 rearrangement, and 1 CBFA2T3-GLIS2 model. Model identities are validated by comparing the PDX STRs, flow cytometry, and fluorescence in situ hybridization results with matched samples from the patient of origin. For 3 models we also have targeted DNA sequencing for 174 recurrent leukemia-associated mutations for both the patient and the corresponding PDX model. All mutations detected in the patient samples were also detected in the PDX models. The Pearson correlation coefficients comparing the variant allele frequencies for each patient-PDX pair range from 0.82 to 0.99, indicating that the models are faithful representations of the original disease even at the subclone level. A summary of the current collection of serially transplanting models is provided in Table 1. Over 15 additional samples have engrafted once and are currently in secondary recipients. Data on established and validated models are being made publicly available through the Baylor PDX Portal (https://pdxportal.research.bcm.edu), where investigators from Baylor and labs worldwide can access information about our collection and request viably frozen samples for their own research purposes. Our models have been distributed to labs around the world for preclinical studies of novel therapeutics.
Conclusion: Through development and distribution of unique pediatric AML PDX models, we hope to create a renewable resource to speed preclinical research and expedite development of drugs for clinical testing for children with AML.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.